The team’s new technique could help scientists worldwide identify novel targets for cancer therapies.
Every writer needs a good editor. That’s true for the publishing business, but it’s also the case for living cells. Our cells create messages using the language of RNA to translate our genetic code into proteins. Those RNA messages are in constant need of editing to help avoid cancer and other diseases. That editing process is crucial for health, but it has always been impossible to observe in close detail — until now.

A team of WashU chemists led by graduate student Alex Quillin has developed a test called EndoVIA that makes it possible to precisely track individual edits in cells, an advance that could lead to a new understanding of the origins of many illnesses. “Our lab is really excited by the development,” Quillin said. “We’ve never been able to pinpoint edited RNA in cells until now.”
The groundbreaking findings were published in the journal ACS Central Science. Co-authors include Jennifer Heemstra, the Charles Allen Thomas Professor of Chemistry, and Benoit Arnould, a postdoctoral research associate.
“RNA editing is critical for our cells to function and often goes awry in disease,” Heemstra said. “However, relative to the tremendous importance of this process, relatively little is known about the timing, location, and regulation of editing in cells.”
The most common edit to RNA is fairly straightforward: Cells use a special enzyme to change molecules of adenosine, a building block of RNA, into another building block, inosine.
This swap, which happens naturally in a wide variety of tissues, is thought to help the body tell the difference between normal RNA and foreign RNA from viruses or other pathogens. “If RNA doesn’t have these edits, the cell can sound the alarm and alert the immune system to take action,” Quillin said.
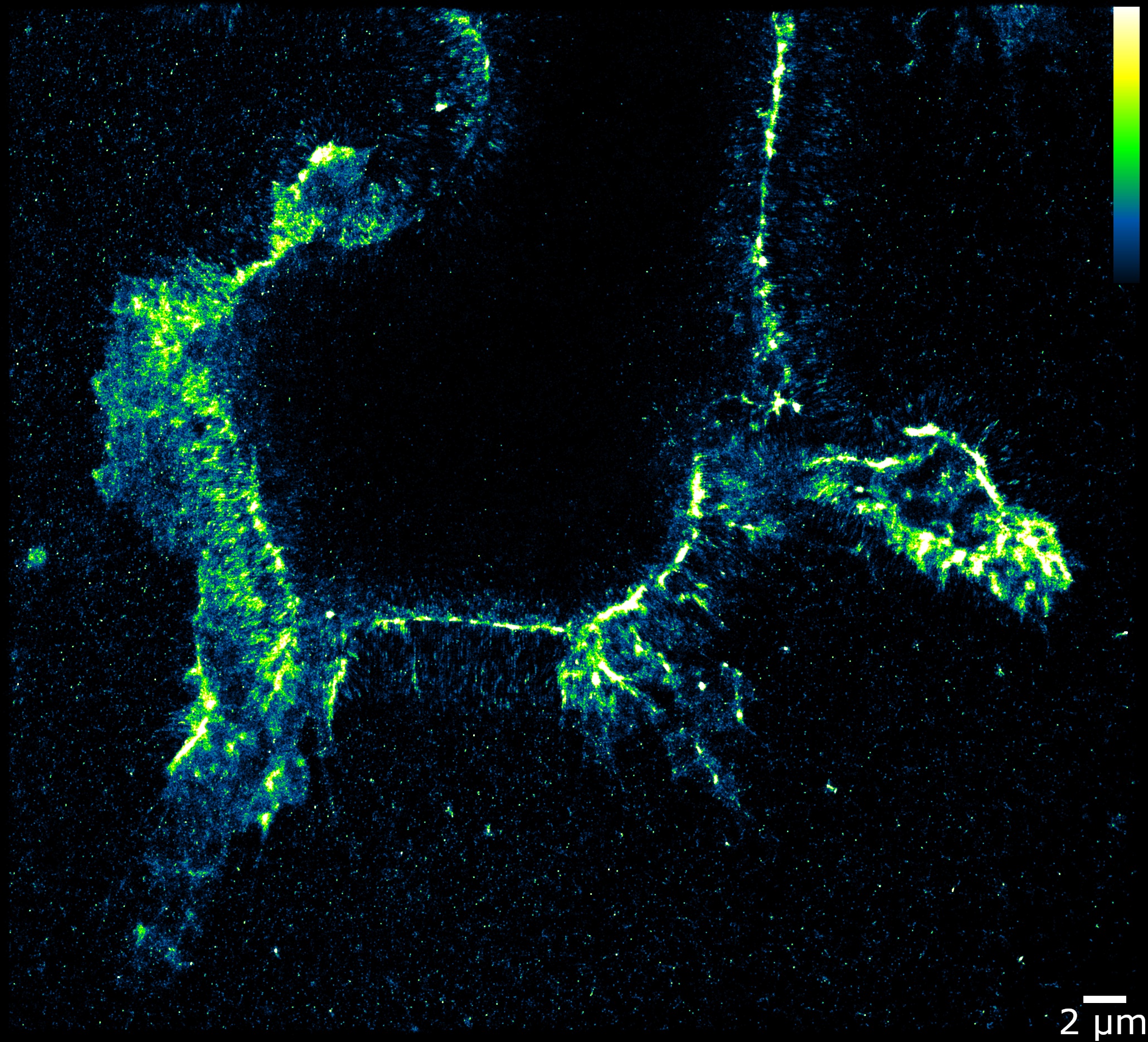
Previous methods for tracking RNA edits required removing RNA from cells, pooling the molecules, and running the sample through a sequencing machine. That process made it possible to count the number of edits, but not to identify where in the cell they occurred.
To better track those critical edits, the WashU team repurposed a different enzyme that targets edited RNAs. Through significant trial and error, they discovered that if they remove magnesium and replace it with calcium, the enzyme will bind to the edited RNA elements in cells. Researchers can then tag the enzyme with fluorescent antibodies that glow under a microscope, offering an unprecedented look at the editing process.

Arnould’s expertise in microscopy made it possible to see individual RNA molecules in cells with unprecedented clarity, Quillin said.
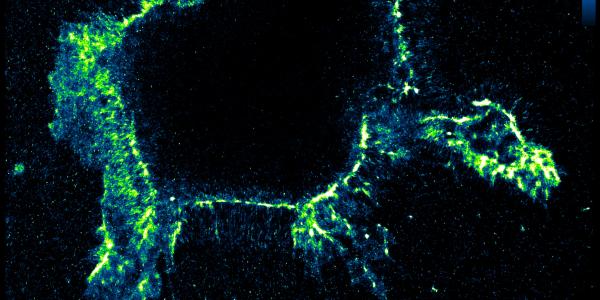
The WashU team’s new technique could help scientists worldwide identify novel targets for cancer therapies, Quillin said. “Some cancers seem to be related to problems with RNA editing,” she said. Breast cancer cells, for instance, tend to show too much editing, while kidney cancer cells often have too little.
In their latest study, Quillin and colleagues examined healthy and cancerous kidney cells. Using EndoVIA, they could see that the healthy cells had large numbers of edits near the cell membranes, modifications missing in the cancerous cells. The meaning of these differences is still unclear, Quillin said, but the finding opens the door for future studies.
“This is the first time anyone has been able to see these localization patterns and so only now can we and others start diving in to understand their biological causes and implications,” Heemstra said. “I’m especially proud of the EndoVIA method and the team of researchers who made it possible.”