Our main research goals are to develop mass-spectrometry-based methods and apply them in collaborations to understand proteins, their solvent accessibility, aggregation, interfaces, affinities of binding, and their folding. We emphasize three general problems: (1) amyloid protein aggregation (e.g., Ab in Alzheimer’s disease), (2) membrane protein conformational changes accompanying interactions with drugs, for example, and (3) viral proteins and their interactions with other proteins and RNA (e.g., Covid, Ebola infections). Additionally, we are extending our methods to understand structural changes in mRNA, motivated by the new mRNA vaccines.
Footprinting gives insight on higher order structure.
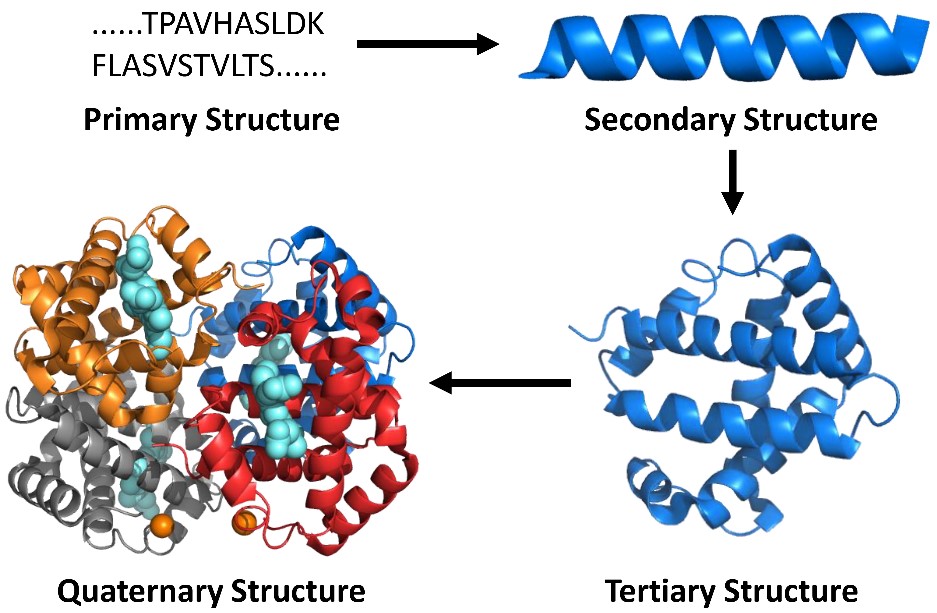
Our subject area is called “structural proteomics”, a new branch of protein science. It builds on proteomics methods, now the principal means of determining primary structure (amino-acid sequence) of proteins, to elucidate secondary, tertiary, and quaternary structure (Figure 1) by “decorating” the proteins with chemical reactions. Although our methods do not afford atomic coordinates, they are well suited to determine changes in structure and conformation that occur upon binding, changes in pH or heating, or mutation. Moreover, they are sensitive and have good throughput.
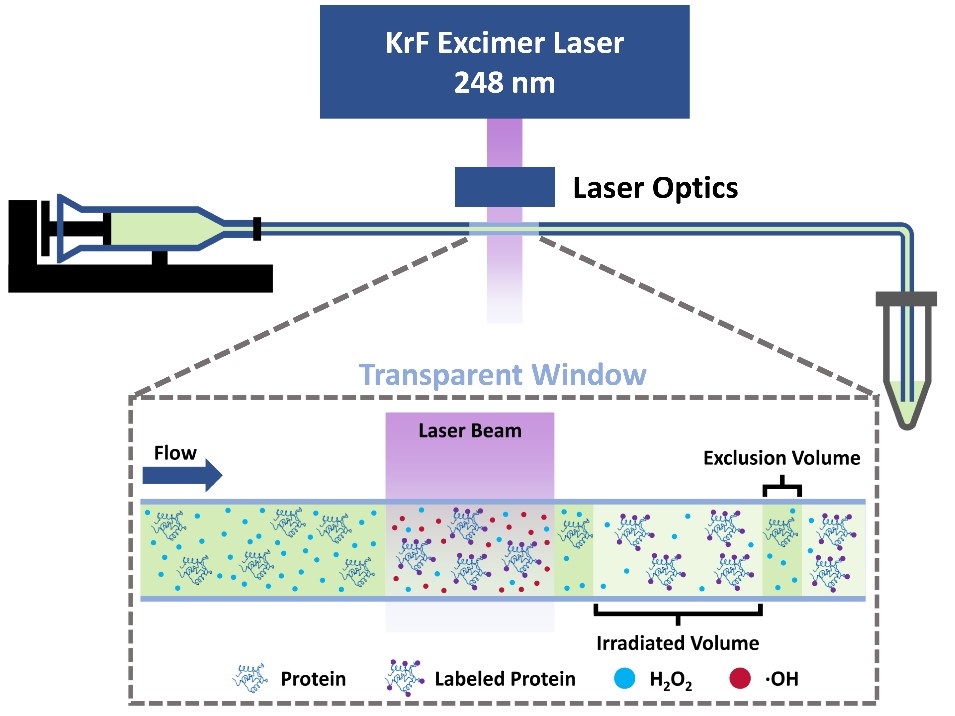
FPOP: One approach for footprinting is to map proteins with reactions of •OH radicals. We termed our version “fast photochemical oxidation of proteins” or FPOP whereby certain surface-accessible amino-acid residues are modified by chemical reactions conducted on the microsecond timescale in a flow tube. The reactive species are generated by pulsed photolysis of H2O2 by a pulsed KrF laser (248 nm). With appropriate scavengers, the reaction time is controlled. FPOP is useful for determining protein-protein interfaces, affinities for tiny amounts of proteins in biophysics, drug discovery, and epitope mapping. FPOP is being adopted by biotechnology companies for quality control of protein therapeutics.
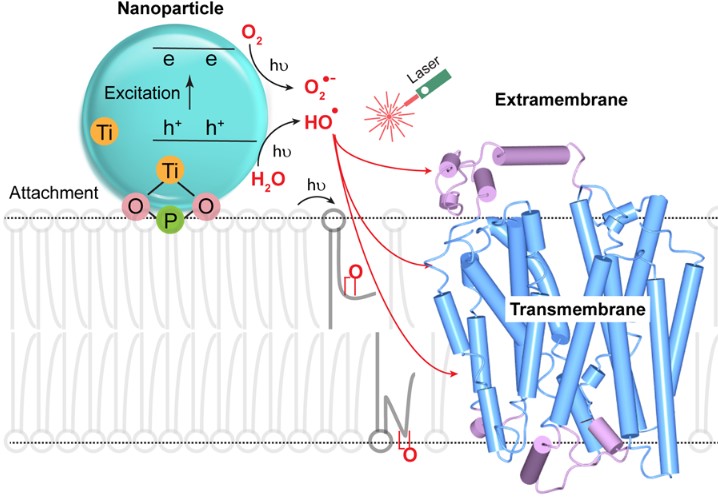
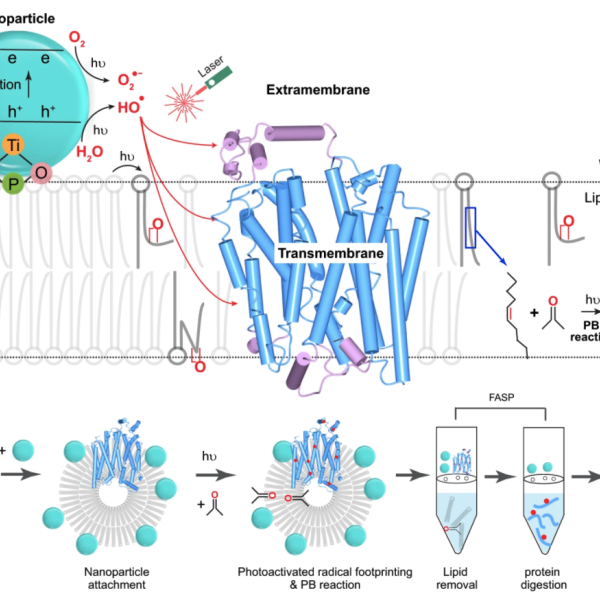
Membrane Proteins: We are extending FPOP to membrane proteins that are difficult to determine by X-ray, NMR, and CryoEM because they are not water soluble and are unstable outside the membrane. Instead of generating the radical from H2O2, we attach a nanoparticle to a liposome (or a living cell). By irradiating it with the FPOP laser, we produce OH radicals adjoining the liposome and admit them to the membrane by perturbing it by a chemical reaction initiated by the footprinting laser. Footprinting can use many other reactions, and we are developing new reactions and new methods and applying these reactions to difficult problems. For example, for fast labeling, we are expanding to carbenes and carbocations. For specific amino-acid footprinting, we use labeling with NEM (N-ethylmaleimide) to follow changes in the local environment of Cys, GEE (glycyl ethyl ester) to monitor changes of Asp and Glu, for example.
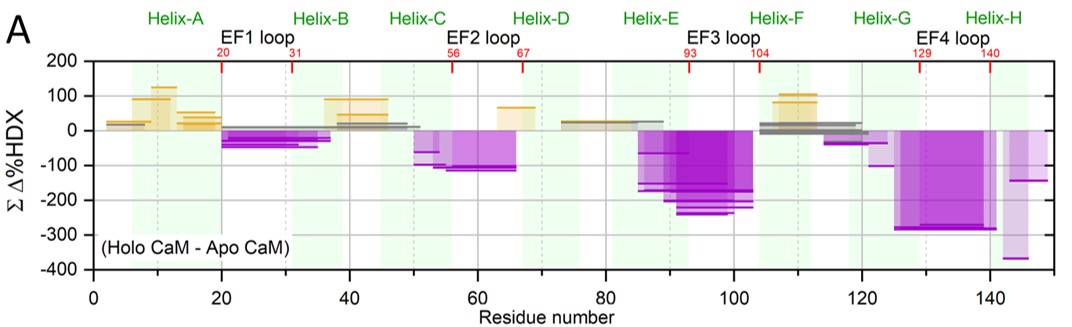
Hydrogen/deuterium exchange (HDX): HDX is another focus where we develop new methods and use them in problem solving. Examples are pulsed HDX for following aggregation of Aβ, the plaque-forming protein in Alzheimer’s disease, and pH-dependent HDX for monitoring protein conformational changes as pH decreases. We also developed an HDX titration platform to determine protein-ligand affinities, a method we call PLIMSTEX (Protein Ligand Interaction by Mass Spectrometry, Titration and H/D Exchange). We wish to apply this to determining affinities for specific regions of proteins, which is difficult by standard measurements. As an example, Figure 4 shows a convincing example of how the protein calmodulin binds Ca2+ to alternately tighten or protect some regions (purple), expose other regions (gold), and have no effect on others (gray).
Crosslinking, Native MS, Ion Mobility: We also utilize chemical cross linking and native electrospray with ion mobility, where near-native forms of a protein or a protein complex are introduced into a mass spectrometer. We discovered that activating the entire complex by electron-capture dissociation (ECD) causes flexible regions of the constituent proteins to fragment. We complement these measurements with native MS, to examine stoichiometry of protein complexes, collisionally induced unfolding, to observe conformational change, and chemical cross linking, to determine topology of the complex.
This research prepares graduate and postdoctoral student to enter academics or biopharma and biotechnology labs, where scores of former coworkers now work.
Examples of applications are ongoing collaborations with
- Carl Frieden and Greg DeKoster (WU Biochemisty and Molecular Biophysics or BMB), Thomas Brett (WU Medicine), and Andrew Yoo (Developmental Biology) on amyloids.
- Weikai Li (BMB) on membrane proteins,
- Gaya Amarasinghe, Daisy Leung, and Daved Fremont (WU Immunology) on viral proteins,
- Leah Wang and Brian Gau (Pfizer) on structural changes of mRNA